About SSBD
What’s SSBD?
SSBD is a platform for sharing and reusing bioimaging data. SSBD consists of a public repository SSBD:repository and an added-value database SSBD:database.
SSBD:repository is a repository service that accepts and shares all kinds of bioimaging data and related data at the time of paper submission or publication. There is no fee for using this service. There is no limit on the data size, but please contact us in advance if the data size exceeds terabytes. We issue a DOI for each project corresponding to your paper. If you wish to share your data, please refer to Share your data.
SSBD:database is an added-value database in which curators select and share highly reusable bioimaging data and quantitative biological dynamics data with rich metadata at the time of and after publication of your paper. Our curators will contact you by email or other methods to ask about data sharing. Please contact us if you would like to share your data in SSBD:database.
SSBD allows users to search, preview, and download shared bioimaging data. For re-analysis and reproduction, downloadable data is as similar as possible to the data as it was obtained by microscopy. Please refer to the Image submission guidelines for our policy when registering bioimaging data. For this reason, data may be shared in the original format from the microscope. SSBD uses the SSBD:OMERO system for previewing microscope images and displaying metadata.
SSBD also provides access to some metadata via a RESTful API.
SSBD allows you to search, visualize, and download shared quantitative biological dynamics data. Please refer to the Quantitative data submission guidelines for our policy when registering quantitative biological dynamics data. For reuse, downloadable data is provided in BDML/BD5 format. SSBD uses the webbd5viewer system for visualization and metadata display of quantitative biological dynamics data. For data in BDML/BD5 format registered with SSBD, we provide XML format retrieval and access via the HDF5 API.
SSBD provides some metadata in RDF. RDF data is also available in the Riken MetaDatabase and the NBDC RDF Portal.
For more information on SSBD, please refer to the following papers:
Koji Kyoda, Hiroya Itoga, Yuki Yamagata, Emi Fujisawa, Fangfang Wang, Miguel Miranda-Miranda, Haruna Yamamoto, Yasue Nakano, Yukako Tohsato, Shuichi Onami,
SSBD: an ecosystem for enhanced sharing and reuse of bioimaging data,
Nucleic Acids Research , Volume 53, Issue D1, 6 January 2025, Pages D1716-D1723,
doi: 10.1093/nar/gkae860, PMID: 39479781; PMCID: PMC11701685.
Yukako Tohsato, Kenneth H. L. Ho, Koji Kyoda, Shuichi Onami,
SSBD: a database of quantitative data of spatiotemporal dynamics of biological phenomena,
Bioinformatics , Volume 32, Issue 22, November 2016, Pages 3471-3479,
doi: 10.1093/bioinformatics/btw417, PMID: 27412095; PMCID: PMC5181557.
For related papers, please refer to the publications.
SSBD metadata
SSBD shares bioimaging data with own metadata.
- SSBD metadata v1 - Based on BDML metadata, each dataset has its own image information, microscopy information, Gene Ontology, etc.
- SSBD metadata v2 - In addition to SSBD metadata v1, v2 defines project / dataset structure, v2 includes project information, more detailed biological image method ontology, Gene Ontology, MeSH term ontology, etc. Gene and protein information is included in some projects and datasets.
- SSBD metadata v3 - In addition to SSBD metadata v2, v3 is REMBI compatibility, v3 includes more detailed gene and protein information, detailed microscopy commercial information, experimental ontology and treatement ontology, etc.
Currently, SSBD:database and SSBD:repository provide metadata based on SSBD metadata v2.
For data collection, we have already started using SSBD metadata v3 and plan to start providing metadata based on SSBD metadata v3.
We are currently developing a metadata database for SSBD metadata v3. A preview version is now available (October 20, 2025). https://ssbd.riken.jp/v3/projects
Please refer to SSBD metadata for the detail.
History
In 2013, The SSBD database was developed and started its operation under the Life Science Database Integration Project National Bioscience Database Center (NBDC), Japan Science and Technology Agency (JST). Initially, as the “Integration of Databases for System Science of Biological Dynamics” project, the SSBD database shared quantitative biological dynamics data and pre-analytical microscopy images.
In 2015, as the “Integrated Database of Biological Dynamics Data and Cellular/Developmental Image Data” project funded by NBDC/JST, we started to share microscopic images without quantitative biological dynamics data, as well as started repository service for to-be-published papers to share microscopic images and quantitative biological dynamics data.
From 2019, the SSBD database has been reorganized into two services: the public repository service SSBD:repository and the added-value database SSBD:database, as a platform for the bioimaging data ecosystem.
From 2022, as the “Establishment of a global data sharing system for bioimaging data” project also funded by Life Science Database Integration Project (Database Integration Coordination Program), Department of NBDC Program, JST, we are working on worldwide metadata sharing and inter-exchange of data, mainly in collaboration with the Image Data Resource (IDR) and the BioImage Archive (BIA) in Europe.
From 2024, the foundingGIDE project, funded by Europe Horizon, is positioned as one of the main platforms for sharing life science and pre-crinical bioimaging data between Japan, Europe and Australia.
Funding
The SSBD has been developed with the support of
- Life Science Database Integration Project (Database Integration Coordination Program), Department of NBDC Program, Japan Science and Technology Agency (JPMJND2201)
- Europe Horizon foundingGIDE Project
- RIKEN
- Japan Science and Technology Agency, Core Research for Evolutional Science and Technology (CREST)
- MEXT, Grant-in-Aid for Scientific Research (KAKENHI)
Collaborations
The SSBD project is collaborating with the following projects to share data and operata data portals.
- CREST Multicellular interaction Development of technology platforms for quantitative analysis of spatiotemporal multicellular interaction project (Data portal)
- Singularity Biology project, Grant-in-Aid for Scientific Research, New Scientific Area (Data portal)
Sister databases
The SSBD is operating the following sister databases. All of them are added-value databases with specific metadata and visualization systems, based on SSBD:repository and SSBD:database as the infrastructure for bioimage data and metadata.
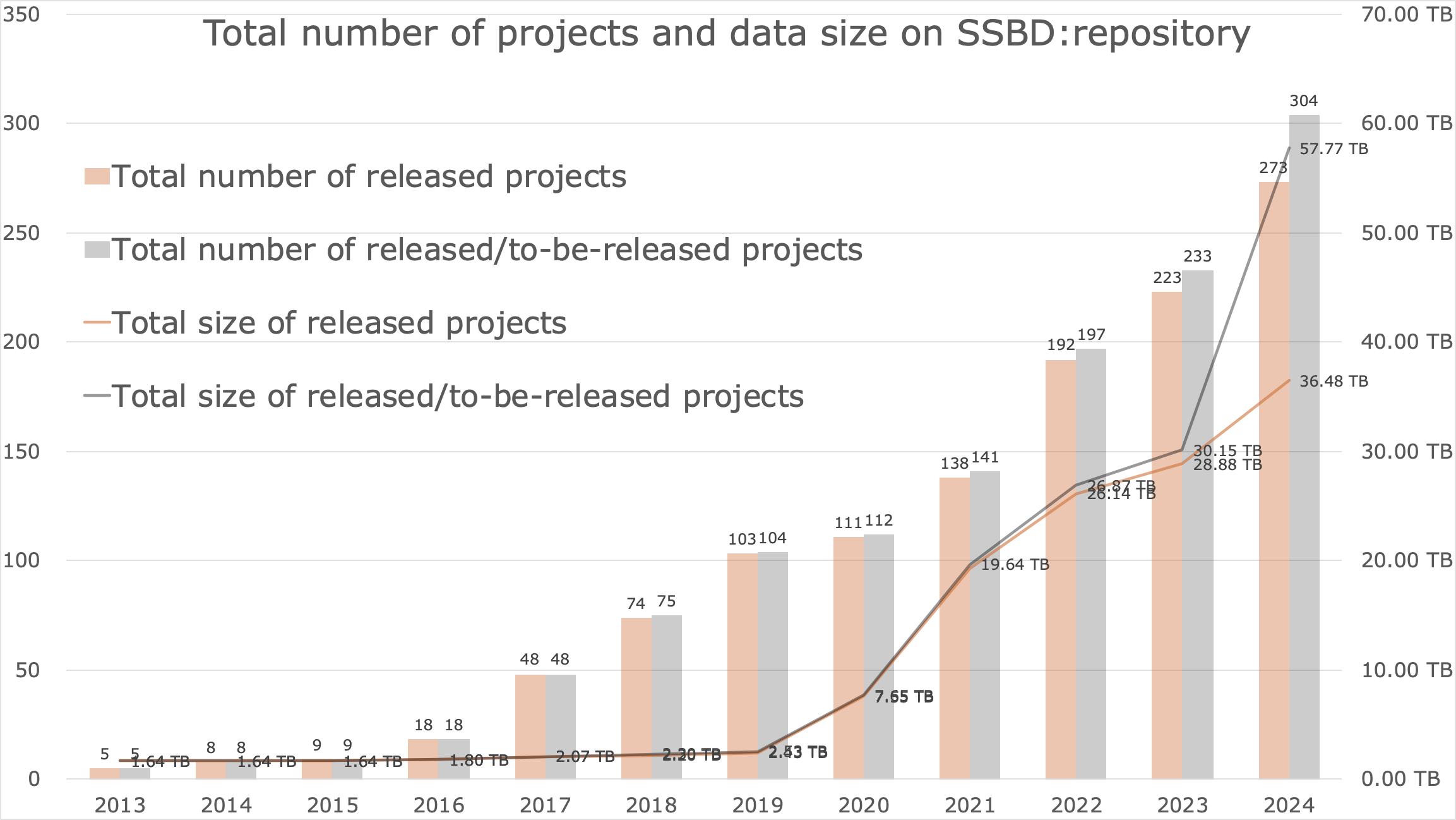
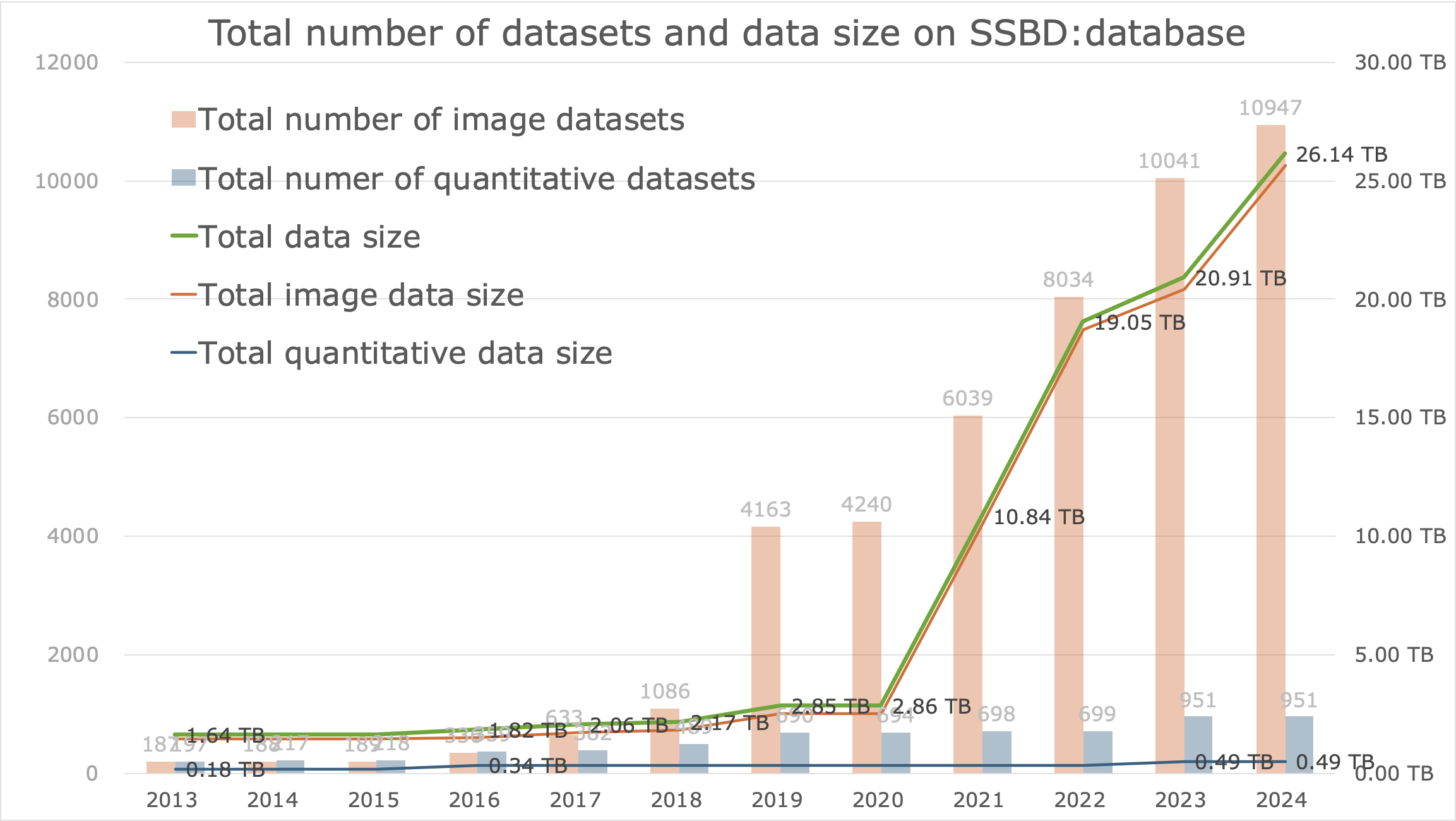
Statistics
(2024-12-24)
OME-NGFF/OME-Zarr
SSBD is committed to the OME-NGFF/OME-Zarr project and has already providing some data in OME-Zarr format. Please visit SSBD OME-NGFF Samples
BDML/BD5
SSBD is providing quantitative biological data in BDML/BD5 format. For more information on BDML/BD5, see BDML/BD5 format